
Data versioning
data_versioning.RmdThis vignette will cover the data versioning functionality of the
resrepo package, considering the following use cases:
- Your data are too large to be stored in a git repository
- You want to explore alternative workflows for your data analysis
- You want to store your data externally
The function version_setup is used in each of these
cases to create a versioned repository to work with. We begin in this
way to maintain consistent structure of resrepo
repositories.
‘Setting up data versioning’ illustrates how to use the function
version_setupto create a versioned repository. This will prevent git from tracking data.‘Adding a new version’ will show you how to explore alternative approaches to data analysis (e.g. a different version of data filtering). This section illustrates how to add new versions of the code and data, and merge branches.
‘Outsourcing the data’ demonstrates show how to use
version_setupto outsource your data storage to a different location, such as a folder synced to the cloud or a hard drive. You can still add new versions and merge branches when outsourcing your data.
Setting up data versioning
Let us start by recreating the example repository. This is the same
repository we created in vignette workflow; if you still
have that repository, you don’t need to run the code below:
library(resrepo)
init_resrepo()
file.copy(
from = system.file("vignette_example/tux_measurements.csv",
package = "resrepo"
),
to = path_resrepo("/data/raw/original/tux_measurements.csv"),
overwrite = TRUE
)
file.copy(
from = system.file("vignette_example/s01_download_penguins.Rmd",
package = "resrepo"
),
to = path_resrepo("/code/s01_download_penguins.Rmd"),
overwrite = TRUE
)
knit_to_results(path_resrepo("/code/s01_download_penguins.Rmd"))
#> processing file: s01_download_penguins.Rmd
#> output file: s01_download_penguins.knit.md
#>
#> Output created: s01_download_penguins.pdf
file.copy(
from = system.file("vignette_example/s02_merge_clean.Rmd",
package = "resrepo"
),
to = path_resrepo("/code/s02_merge_clean.Rmd"),
overwrite = TRUE
)
knit_to_results(path_resrepo("/code/s02_merge_clean.Rmd"))
#> processing file: s02_merge_clean.Rmd
#> output file: s02_merge_clean.knit.md
#>
#> Output created: s02_merge_clean.pdf
file.copy(
from = system.file("vignette_example/s03_pca.Rmd", package = "resrepo"),
to = path_resrepo("/code/s03_pca.Rmd"),
overwrite = TRUE
)
knit_to_results(path_resrepo("/code/s03_pca.Rmd"))
#> processing file: s03_pca.Rmd
#> output file: s03_pca.knit.md
#>
#> Output created: s03_pca.pdf
git2r::add(path = ".")
git2r::commit(message = "Set up", all = TRUE)Let us check that we do have indeed a full repository:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ ├── s01_download_penguins.Rmd
#> │ ├── s02_merge_clean.Rmd
#> │ └── s03_pca.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean
#> │ │ ├── penguins_na_omit.csv
#> │ │ ├── renv_s02_merge_clean.json
#> │ │ └── renv_s03_pca.json
#> │ └── raw
#> │ ├── README.md
#> │ ├── original
#> │ │ └── tux_measurements.csv
#> │ └── s01_download_penguins
#> │ ├── palmer_penguins.csv
#> │ └── renv_s01_download_penguins.json
#> ├── results
#> │ ├── README.md
#> │ ├── s01_download_penguins
#> │ │ ├── README.md
#> │ │ └── s01_download_penguins.pdf
#> │ ├── s02_merge_clean
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean.pdf
#> │ └── s03_pca
#> │ ├── README.md
#> │ ├── s03_pca.pdf
#> │ └── s03_pca_files
#> │ └── figure-latex
#> │ └── pca_plot-1.png
#> └── writing
#> └── README.mdWhen we want to start versioning the data, we can use the function
version_setup. We only need to use the function once at the
beginning of the project to initialise versioning. You can specify the
location of your data using the resources_path argument.
The function will create a subfolder named versions in this
location.
By default, this will be stored in your repository, but it doesn’t have to, and it can be located anywhere on your system (see the second part of this vignette for an example).
version_setup(quiet = TRUE, resources_path = NULL)
#> [1] TRUELet us check what happened:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ ├── s01_download_penguins.Rmd
#> │ ├── s02_merge_clean.Rmd
#> │ └── s03_pca.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ ├── raw
#> │ └── version_meta
#> │ ├── initial.meta
#> │ ├── intermediate_in_use.meta
#> │ ├── raw_in_use.meta
#> │ └── starting.meta
#> ├── results
#> │ ├── README.md
#> │ ├── s01_download_penguins
#> │ │ ├── README.md
#> │ │ └── s01_download_penguins.pdf
#> │ ├── s02_merge_clean
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean.pdf
#> │ └── s03_pca
#> │ ├── README.md
#> │ ├── s03_pca.pdf
#> │ └── s03_pca_files
#> │ └── figure-latex
#> │ └── pca_plot-1.png
#> ├── versions
#> │ ├── initial
#> │ │ └── intermediate
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean
#> │ │ ├── penguins_na_omit.csv
#> │ │ ├── renv_s02_merge_clean.json
#> │ │ └── renv_s03_pca.json
#> │ └── starting
#> │ └── raw
#> │ ├── README.md
#> │ ├── original
#> │ │ └── tux_measurements.csv
#> │ └── s01_download_penguins
#> │ ├── palmer_penguins.csv
#> │ └── renv_s01_download_penguins.json
#> └── writing
#> └── README.mdWe can see that both data/raw and
data/intermediate have been moved into
versions, intermediate is under the
initial version, while raw is under the
starting directory. In this vignette, we will be focusing
only on versioning data/intermediate. Advanced users can
also version data/raw (see the end of this vignette). For
now, raw_in_use.meta will point to the starting version of
data/raw and will not change.
data/intermediate and data/raw have now
become links, which is why they are highlighted in a different colour
(light purple) in the tree above. We can use these links as if they were
directories, and check that they point correctly to the directories in
versions.
dir(path_resrepo("/data/raw"))
#> [1] "original" "README.md" "s01_download_penguins"However, note that the data are now NOT being tracked by git, as
versions is in .gitignore. So, you want to make sure that
you have a reliable way to back up your data.
Adding a new version
We might now want to revise the filtering of data that we are doing.
This will affect some intermediate data and results. Whilst results are
tracked in the git repository, data are not. However, we might want to
compare the results of the alternative filtering approaches. We can use
the version_add() function to create a new version of the
data, which we can modify and compare to the initial
version we created earlier. It will also create a new branch in the git
repository (with the same name as the data version):
version_add(
intermediate_new_version = "new_filtering",
intermediate_description = "Filtering out some data"
)
#> version new_filtering created
#> [1] TRUEWe have called our new version new_filtering, and we can
now see that we have switched to a new branch in the git repository
called new_filtering, as indicated by (HEAD) below:
git2r::branches()
#> $main
#> [5e0def] (Local) main
#>
#> $new_filtering
#> [363786] (Local) (HEAD) new_filteringNow data/raw and data/intermediate are
symlinks to the new_filtering version of the data, which is
found under versions/new_filtering. Each version will be
under a new branch of the repository.
It is also in theory possible to version data/raw by
adding “raw_new_version” to the version_add function, but
this is NOT RECOMMENDED, as raw data should ideally not be changed (it
is supposed to represent the data in their original format).
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ ├── s01_download_penguins.Rmd
#> │ ├── s02_merge_clean.Rmd
#> │ └── s03_pca.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ ├── raw
#> │ └── version_meta
#> │ ├── initial.meta
#> │ ├── intermediate_in_use.meta
#> │ ├── new_filtering.meta
#> │ ├── raw_in_use.meta
#> │ └── starting.meta
#> ├── results
#> │ ├── README.md
#> │ ├── s01_download_penguins
#> │ │ ├── README.md
#> │ │ └── s01_download_penguins.pdf
#> │ ├── s02_merge_clean
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean.pdf
#> │ └── s03_pca
#> │ ├── README.md
#> │ ├── s03_pca.pdf
#> │ └── s03_pca_files
#> │ └── figure-latex
#> │ └── pca_plot-1.png
#> ├── versions
#> │ ├── initial
#> │ │ └── intermediate
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean
#> │ │ ├── penguins_na_omit.csv
#> │ │ ├── renv_s02_merge_clean.json
#> │ │ └── renv_s03_pca.json
#> │ ├── new_filtering
#> │ │ └── intermediate
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean
#> │ │ ├── penguins_na_omit.csv
#> │ │ ├── renv_s02_merge_clean.json
#> │ │ └── renv_s03_pca.json
#> │ └── starting
#> │ └── raw
#> │ ├── README.md
#> │ ├── original
#> │ │ └── tux_measurements.csv
#> │ └── s01_download_penguins
#> │ ├── palmer_penguins.csv
#> │ └── renv_s01_download_penguins.json
#> └── writing
#> └── README.mdImagine that we have just been told that the first 10 measurements of
the penguins might be considered unreliable, as the scales were showing
a low battery warning. We now will edit the code that merges and cleans
the data, and modify it to remove the first 10 penguins from the
dataset. We use a pre-prepared script for this, which we copy to the
code directory:
file.copy(
from = system.file("vignette_example/new_filtering/s02_merge_clean.Rmd",
package = "resrepo"
),
to = path_resrepo("code/s02_merge_clean.Rmd"),
overwrite = TRUE
)
#> [1] TRUEIf this was a “real” project, since the new_filtering
version is a new branch of the repository, we could edit the script in
the code directory and commit the changes.
We can now remove the full version of the intermediate dataset in the
new_filtering folder so that we can replace it with a new
version that excludes the first 10 penguins.
unlink(path_resrepo("data/intermediate/s02_merge_clean"), recursive = TRUE)Note that penguins_na_omit.csv is still in the
versions/initial folder, as we have not removed it from
versions/initial but only from the
versions/new_filtering directory.
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ ├── s01_download_penguins.Rmd
#> │ ├── s02_merge_clean.Rmd
#> │ └── s03_pca.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ ├── raw
#> │ └── version_meta
#> │ ├── initial.meta
#> │ ├── intermediate_in_use.meta
#> │ ├── new_filtering.meta
#> │ ├── raw_in_use.meta
#> │ └── starting.meta
#> ├── results
#> │ ├── README.md
#> │ ├── s01_download_penguins
#> │ │ ├── README.md
#> │ │ └── s01_download_penguins.pdf
#> │ ├── s02_merge_clean
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean.pdf
#> │ └── s03_pca
#> │ ├── README.md
#> │ ├── s03_pca.pdf
#> │ └── s03_pca_files
#> │ └── figure-latex
#> │ └── pca_plot-1.png
#> ├── versions
#> │ ├── initial
#> │ │ └── intermediate
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean
#> │ │ ├── penguins_na_omit.csv
#> │ │ ├── renv_s02_merge_clean.json
#> │ │ └── renv_s03_pca.json
#> │ ├── new_filtering
#> │ │ └── intermediate
#> │ │ └── README.md
#> │ └── starting
#> │ └── raw
#> │ ├── README.md
#> │ ├── original
#> │ │ └── tux_measurements.csv
#> │ └── s01_download_penguins
#> │ ├── palmer_penguins.csv
#> │ └── renv_s01_download_penguins.json
#> └── writing
#> └── README.mdIf we now knit the new version of the code, where we remove the first
10 penguins, the data are written to the
versions/new_filtering/intermediate directory.
knit_to_results(path_resrepo("/code/s02_merge_clean.Rmd"))
#> processing file: s02_merge_clean.Rmd
#> output file: s02_merge_clean.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS s02_merge_clean.knit.md --to latex --from markdown+autolink_bare_uris+tex_math_single_backslash --output s02_merge_clean.tex --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --highlight-style tango --pdf-engine pdflatex --variable graphics --variable 'geometry:margin=1in'
#>
#> Output created: s02_merge_clean.pdf
#> [1] TRUE
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ ├── s01_download_penguins.Rmd
#> │ ├── s02_merge_clean.Rmd
#> │ └── s03_pca.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ ├── raw
#> │ └── version_meta
#> │ ├── initial.meta
#> │ ├── intermediate_in_use.meta
#> │ ├── new_filtering.meta
#> │ ├── raw_in_use.meta
#> │ └── starting.meta
#> ├── results
#> │ ├── README.md
#> │ ├── s01_download_penguins
#> │ │ ├── README.md
#> │ │ └── s01_download_penguins.pdf
#> │ ├── s02_merge_clean
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean.pdf
#> │ └── s03_pca
#> │ ├── README.md
#> │ ├── s03_pca.pdf
#> │ └── s03_pca_files
#> │ └── figure-latex
#> │ └── pca_plot-1.png
#> ├── versions
#> │ ├── initial
#> │ │ └── intermediate
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean
#> │ │ ├── penguins_na_omit.csv
#> │ │ ├── renv_s02_merge_clean.json
#> │ │ └── renv_s03_pca.json
#> │ ├── new_filtering
#> │ │ └── intermediate
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean
#> │ │ ├── penguins_na_omit.csv
#> │ │ └── renv_s02_merge_clean.json
#> │ └── starting
#> │ └── raw
#> │ ├── README.md
#> │ ├── original
#> │ │ └── tux_measurements.csv
#> │ └── s01_download_penguins
#> │ ├── palmer_penguins.csv
#> │ └── renv_s01_download_penguins.json
#> └── writing
#> └── README.mdWe can now compare the datasets of our two versions, and we can see
that our current version of the data, which is found in
new_filtering, has 10 fewer penguins than the
initial version.
penguins_new_filtering <- read.csv(path_resrepo(
"data/intermediate/s02_merge_clean/penguins_na_omit.csv"
))
penguins_initial <- read.csv(path_resrepo(
"/versions/initial/intermediate/s02_merge_clean/penguins_na_omit.csv"
))
nrow(penguins_initial)
#> [1] 377
nrow(penguins_new_filtering)
#> [1] 367Merging back into main
If we are happy with the new version, we can merge it back into the
main branch. This will mean that our branch main will now
use the new version of the data (with the 10 less penguins). At the same
time, this means that we will replace the previous code with the code in
the new branch. You may want to keep both versions of the code if you
are interested in using both versions of the data. If we are absolutely
sure about merging back into the main branch, before doing it, we need
to commit the changes to the branch new_filtering.
How to commit in RStudio:
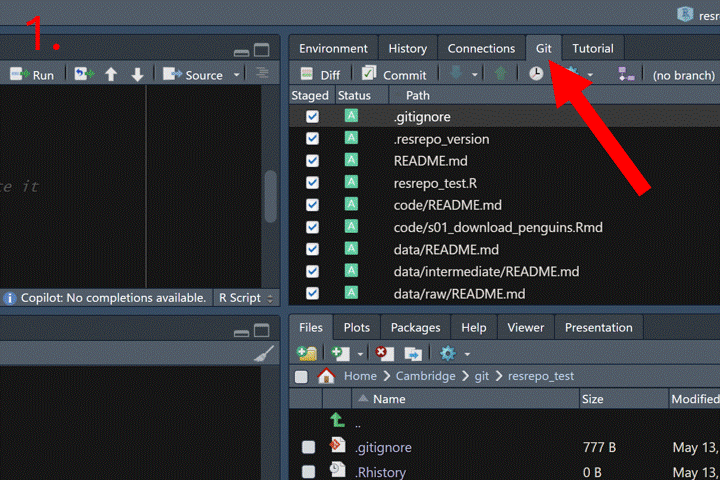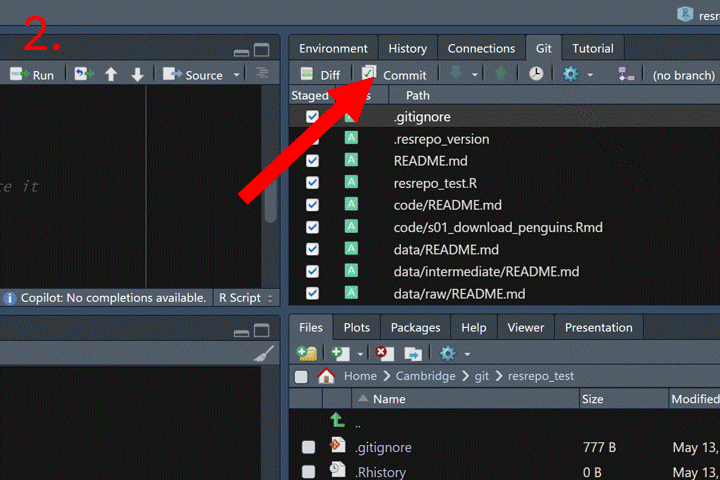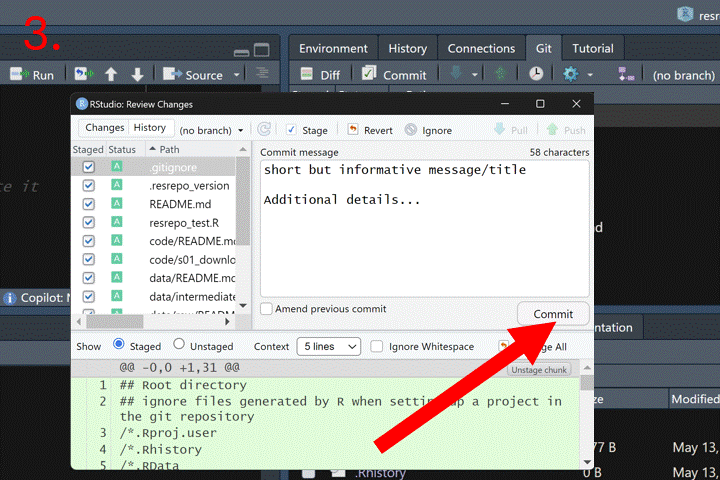
Now we can merge the branch new_filtering into the
branch main, and check out main. To to this,
we can use the GitHub interface, following the steps below.
First, we need to push both branches to GitHub:
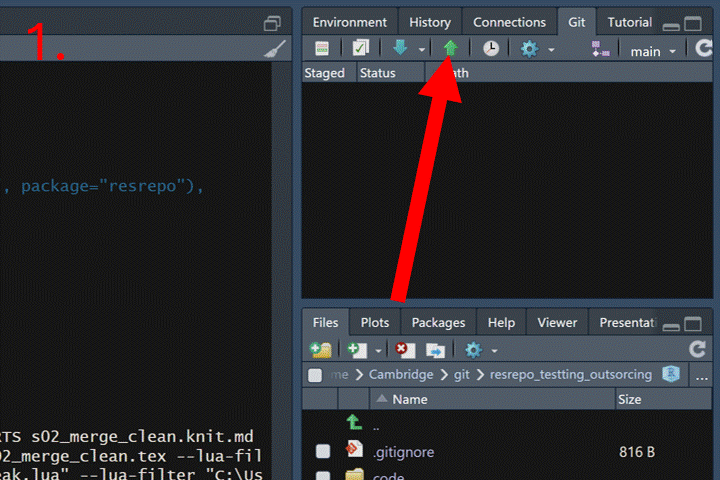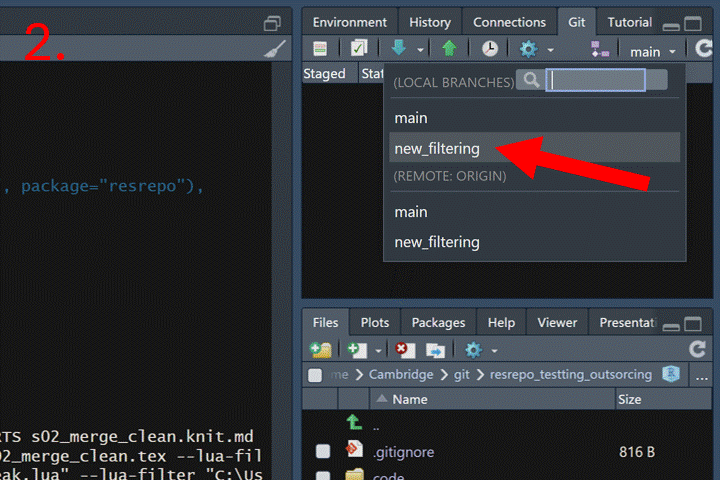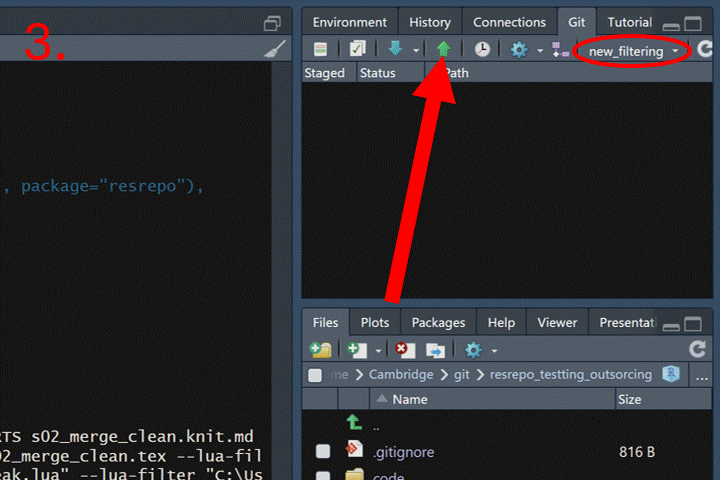
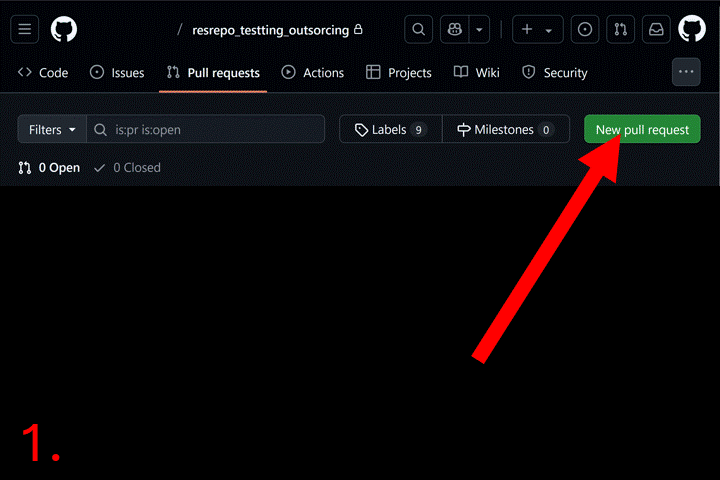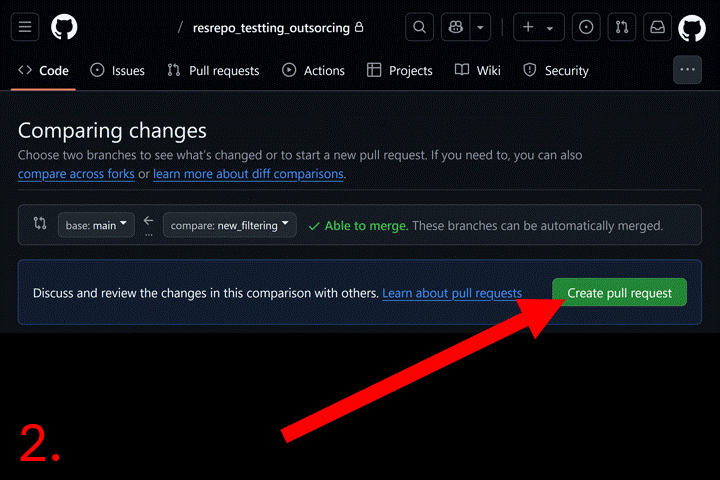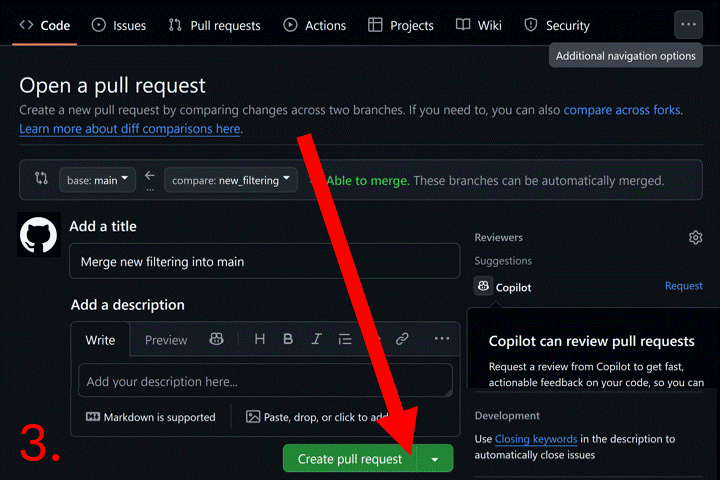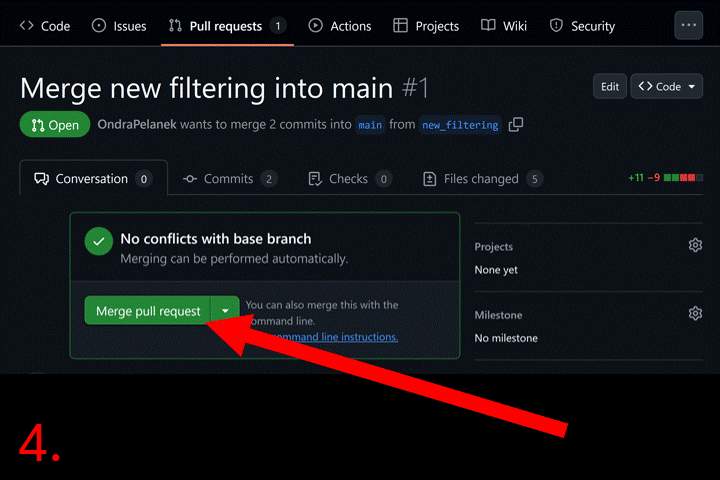
Now go to your repository page on GitHub. Click the “Pull requests”
tab. Then click the “New pull request” button and select base branch
(e.g., main) and compare branch (e.g.,
new_filtering). Click “Create pull request”, and add a
description. Click “Merge pull request” to complete the merge. Now you
can delete the new_filtering branch.
And now we have successfully versioned and updated our repository.
Outsourcing the data
Often, data are too big to be stored within your git repository.
Therefore, we may want to store them into a different location: locally
within your machine, in a hard drive, or to a folder synced to the
cloud. In this section, we will show how to use the
version_setup function to outsource your data.
Be aware that we can use the version_setup function only
once when initializing a repository. Therefore, we need to create an
empty repository for this example:
We now need to create a folder for data storage in our desired external location.
external_data_storage <- file.path(tempdir(), "external_data_storage")
unlink(external_data_storage, recursive = TRUE)
dir.create(external_data_storage)For the purpose of this vignette, the above folder is created in a temporary location. For your project this could be a directory in your local machine, on a hard drive, or a folder synced to the cloud.
Now we have an empty folder where we want to store our data, and an empty repository for our project.
This time to set up versioning with external data storage, we will
again use the function version_setup, however, we will set
the argument resources_path to the external path where we
would like to store the new versions of our data.
version_setup(quiet = TRUE, resources_path = external_data_storage)
#> [1] TRUEWe can now check that the repository has been set up correctly and we
see that external_data_storage is not found in the tree of
our repository:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ └── README.md
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ ├── raw
#> │ └── version_meta
#> │ ├── initial.meta
#> │ ├── intermediate_in_use.meta
#> │ ├── raw_in_use.meta
#> │ └── starting.meta
#> ├── results
#> │ └── README.md
#> ├── versions
#> └── writing
#> └── README.mdHowever, the data/raw and data/intermediate
directories are now symlinks to the external_data_storage
folder, which is where the versions of our data will be stored.
We can see the structure of our external data storage folder looks like this:
fs::dir_tree("../../external_data_storage")
#> ../../external_data_storage
#> └── versions
#> ├── initial
#> │ └── intermediate
#> │ └── README.md
#> └── starting
#> └── raw
#> ├── README.md
#> └── originalWe can now populate our repository with the necessary scripts.
file.copy(
from = system.file("vignette_example/s01_download_penguins.Rmd",
package = "resrepo"
),
to = path_resrepo("/code/s01_download_penguins.Rmd"),
overwrite = TRUE
)
#> [1] TRUE
file.copy(
from = system.file("vignette_example/s02_merge_clean.Rmd",
package = "resrepo"
),
to = path_resrepo("/code/s02_merge_clean.Rmd"),
overwrite = TRUE
)
#> [1] TRUE
file.copy(
from = system.file("vignette_example/s03_pca.Rmd", package = "resrepo"),
to = path_resrepo("/code/s03_pca.Rmd"),
overwrite = TRUE
)
#> [1] TRUEAnd add our data:
file.copy(
from = system.file("vignette_example/tux_measurements.csv",
package = "resrepo"
),
to = path_resrepo("/data/raw/original/tux_measurements.csv"),
overwrite = TRUE
)
#> [1] TRUEWe can now run our scripts:
knit_to_results(path_resrepo("/code/s01_download_penguins.Rmd"))
#> processing file: s01_download_penguins.Rmd
#> output file: s01_download_penguins.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS s01_download_penguins.knit.md --to latex --from markdown+autolink_bare_uris+tex_math_single_backslash --output s01_download_penguins.tex --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --highlight-style tango --pdf-engine pdflatex --variable graphics --variable 'geometry:margin=1in'
#>
#> Output created: s01_download_penguins.pdf
#> [1] TRUE
knit_to_results(path_resrepo("/code/s02_merge_clean.Rmd"))
#> processing file: s02_merge_clean.Rmd
#> output file: s02_merge_clean.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS s02_merge_clean.knit.md --to latex --from markdown+autolink_bare_uris+tex_math_single_backslash --output s02_merge_clean.tex --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --highlight-style tango --pdf-engine pdflatex --variable graphics --variable 'geometry:margin=1in'
#>
#> Output created: s02_merge_clean.pdf
#> [1] TRUE
knit_to_results(path_resrepo("/code/s03_pca.Rmd"))
#> processing file: s03_pca.Rmd
#> output file: s03_pca.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS s03_pca.knit.md --to latex --from markdown+autolink_bare_uris+tex_math_single_backslash --output s03_pca.tex --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --highlight-style tango --pdf-engine pdflatex --variable graphics --variable 'geometry:margin=1in'
#>
#> Output created: s03_pca.pdf
#> [1] TRUEWhen we view the git repository we can now see all our results files:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ ├── s01_download_penguins.Rmd
#> │ ├── s02_merge_clean.Rmd
#> │ └── s03_pca.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ ├── raw
#> │ └── version_meta
#> │ ├── initial.meta
#> │ ├── intermediate_in_use.meta
#> │ ├── raw_in_use.meta
#> │ └── starting.meta
#> ├── results
#> │ ├── README.md
#> │ ├── s01_download_penguins
#> │ │ ├── README.md
#> │ │ └── s01_download_penguins.pdf
#> │ ├── s02_merge_clean
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean.pdf
#> │ └── s03_pca
#> │ ├── README.md
#> │ ├── s03_pca.pdf
#> │ └── s03_pca_files
#> │ └── figure-latex
#> │ └── pca_plot-1.png
#> ├── versions
#> └── writing
#> └── README.mdAnd the data are successfully stored in our external folder:
fs::dir_tree("../../external_data_storage")
#> ../../external_data_storage
#> └── versions
#> ├── initial
#> │ └── intermediate
#> │ ├── README.md
#> │ └── s02_merge_clean
#> │ ├── penguins_na_omit.csv
#> │ ├── renv_s02_merge_clean.json
#> │ └── renv_s03_pca.json
#> └── starting
#> └── raw
#> ├── README.md
#> ├── original
#> │ └── tux_measurements.csv
#> └── s01_download_penguins
#> ├── palmer_penguins.csv
#> └── renv_s01_download_penguins.json