
resrepo
resrepo.Rmdresrepo: easy research on git
The aim of resrepo is to encourage and facilitate good
practices when setting up and managing git repositories for
scientific research projects. There are three main elements to this:
- it provides a template for a tidy repository structure that can be used for any project, with functions that help keeping a clear separation of code, data and results.
- it provides functionality to help manage large data that cannot be tracked on a git repository. This is achieved through versioning the data and results with the use of symlinks, allowing easy comparison of different versions (e.g. different data filtering) and enabling the storage data outside the git repository, potentially in directories that are backed up on the cloud.
General overview
In resrespo, we keep data, code, results, and the
write-up separate. This allows for a tidy structure that can flexibly
accommodate very complex projects. Note that there are a number of
README.md files, which provide instructions and advice related to the
files that go into that particular directory.
If you created the repository with init_resrepo, the
function will automatically commit these changes for you, so that you
have a clean repository template as your first commit, and you can best
document the changes that you make as you progress.
resrepo template
The first way in which resrepo can facilitate good practices when managing git repositories, is to structure your folders so each type of file has their specific location. This helps to separate your data from your code (which is good coding practice) and creates an easy to navigate file structure for all your output.
Here is a general overview of the structure:
- code: this is, unsurprisingly, the folder for your code, which may be in R, or other languages if needed.
-
data:
resrepowill automatically create a data folder that contains two subfolders: raw is intended for your raw/initial set of data, needed to start running the analyses. intermediate, instead, is where most outputs should go (e.g., modifications of the raw data, tables, plots, intermediate input files). Because these folder may end up including huge files, by default the content of “data” will not be uploaded to GitHub, as it is automatically added to.gitignore. - results: this folder should only be reserved for final files (e.g, summary tables, plots) that are intended for publication. Because this folder is intended for a limited number of small files, it is expected to be uploaded on GitHub.
- writing: this folder is intended for the write-up of your project. It can contain markdown files, or links to GoogleDocs or Word documents. It is not recommended to store large files in this folder.
Naming your code scripts
The names of your scripts should be informative and follow a logical
order. We recommend that you use a prefix to indicate the order of the
scripts, such as s01, s02, etc., with an
underscore separating a short but descriptive title. Avoid spaces (they
should be replaced with underscores) and capital letters (not all file
systems/languages are case sensitive). For example, a script that
downloads remote sensing data from a public server could be named
s01_download_remote_sense.Rmd. A script to run a clustering
analysis from the data could be named s02_clustering.Rmd.
Avoid naming your files starting with a number (e.g. instead of
“01_process_data.R” use “s01_process_data.R”) Do not use generic names
such as “Figure_01”, “Figure_02”, etc. for your files and folders. Use
more descriptive names as numbers change. It will make your life easier
when you will need to work again on the project.
Set up data directories
For most projects, you will need some data for your scripts to work. There are two main categories of data: “raw” data, which are primary data (e.g. measurements you made in the lab or the field, FASTQ files generated by a sequencer, remote sensing data downloaded from NASA); and “intermediate” data (data that you generated from raw, and that in turn will be used as the base of further analysis).
In resrepo, we store data in one or more sub-directories
within /data/raw or /data/intermediate,
depending on their category. The template has a
/raw/original subdir, which you could use if you have a
simple project with relatively few data files, but you are free to
remove it and create alternative ones. For data that you collected or
generated, you can use any name (e.g. data/raw/fastq,
data/raw/focal_obs, etc.). If data are generated by a
script, then they should be placed in a subdirectory with the same name
of that script, implicitly documenting their provenance. So, if you
downloaded data from a remote server in a script called
s01_download_remote_sense.Rmd, the data should be stored in
/data/raw/s01_download_remote_sense. If you generated data
in a script called s02_clustering.Rmd, the data should be
stored in /data/intermediate/s02_clustering. Feel free to
have further subdirectories within these directories if you have a large
amount of data.
No files should be stored directly in /data/raw or
/data/intermediate, they should always be put in a first
level sub-directory. If you are not generating any intermediate data,
you can simply ignore the /data/intermediate directory.
Generating results
Some analyses will generate outputs that we want to use in the paper
(e.g. plots or tables). We suggest that you put those in the
results directory, to separate them from intermediate data
(which are generally used for further analysis). As with data, we
suggest that you create a subdirectory for each script that generates
results. So, if you have a script s02_clustering.Rmd that
generates a plot, you should save that plot in
/results/s02_clustering. In this example, the same script
could generate both a large amount of data describing the membership to
clusters (which we decided to store as data) and a plot (which belongs
in results). There are many instances where the same output could be
thought as either data or results; don’t worry too much about it, as
long as you annotate your code well, you will be able to find where the
information is.
Writing it all up
Git is not really designed to handle Word or OpenOffice files.
Ideally, text is kept in markdown files, but that format does not lend
itself nicely to formatting for submission (e.g. bibliography from a
reference manager). Having said that, you could, in principle, track
Word/OpenOffice documents in the writing directory, as they
are rarely large enough to cause trouble. If you are writing
collaboratively, you are more likely to use GoogleDoc or shared Word
documents. In that case, you can use the README.md in the
writing directory to store the paths to those documents. By
doing so, it means that a collaborator who has access to the repository
can also find the manuscript easily (i.e. everything is in one
place).
Initialise the repository
Start by creating a blank repository on GitHub (or equivalent server), and clone it on your computer. Instructions to do so can be found at this link or you can follow the gif below. For this vignette, we will create a “resrepo_example” git repository.
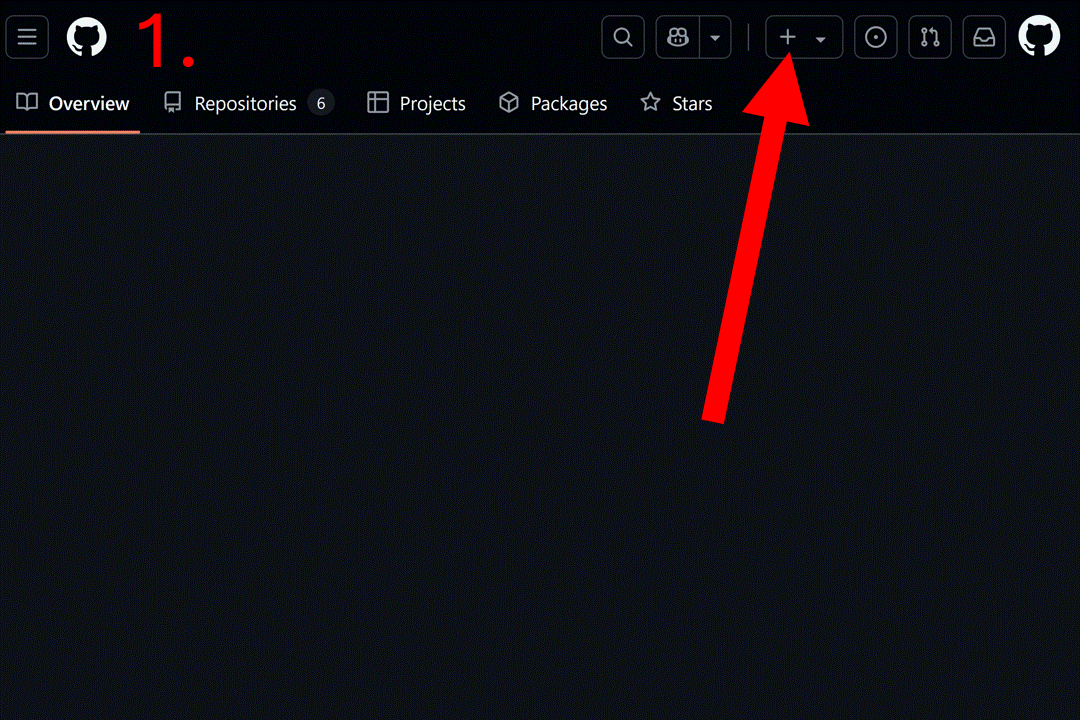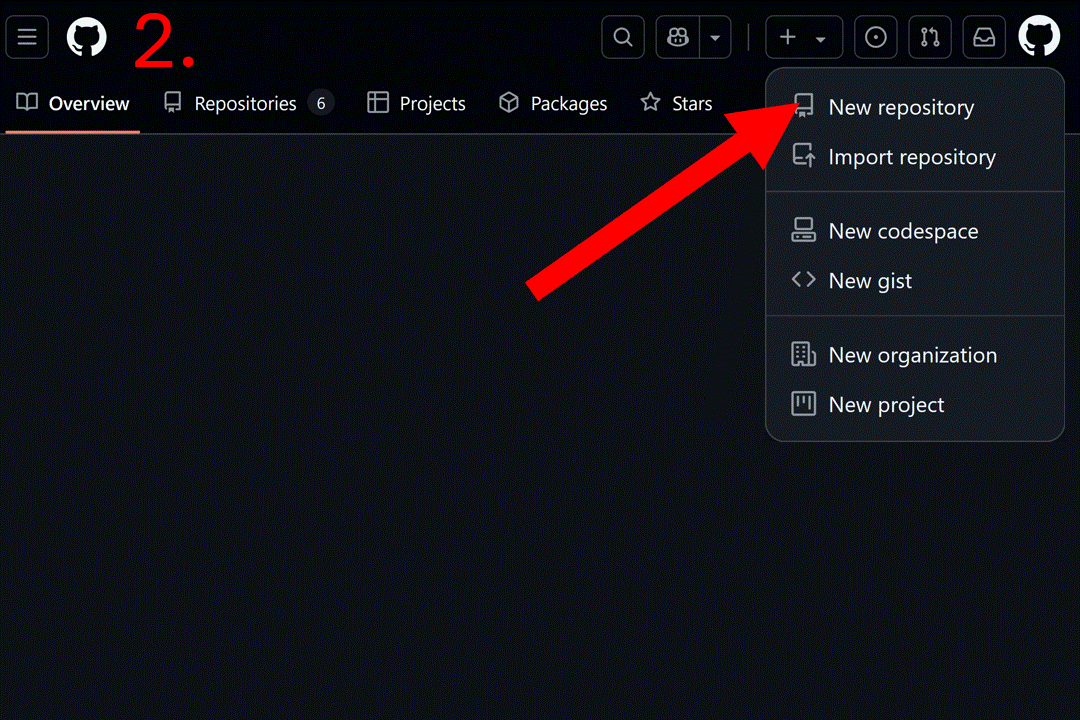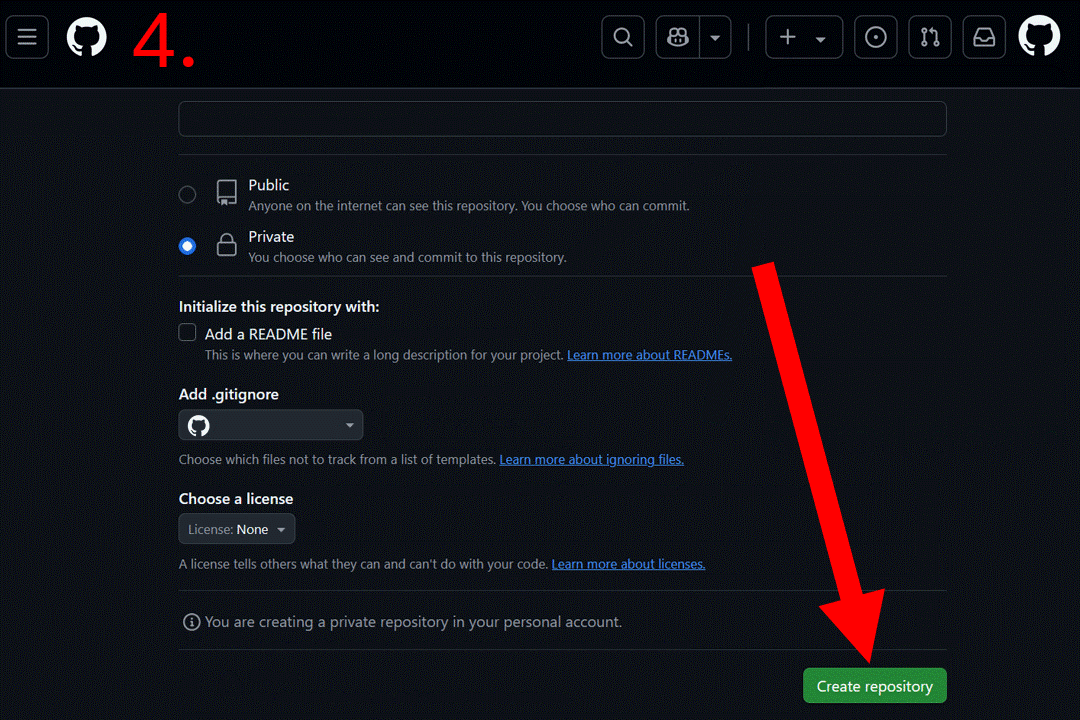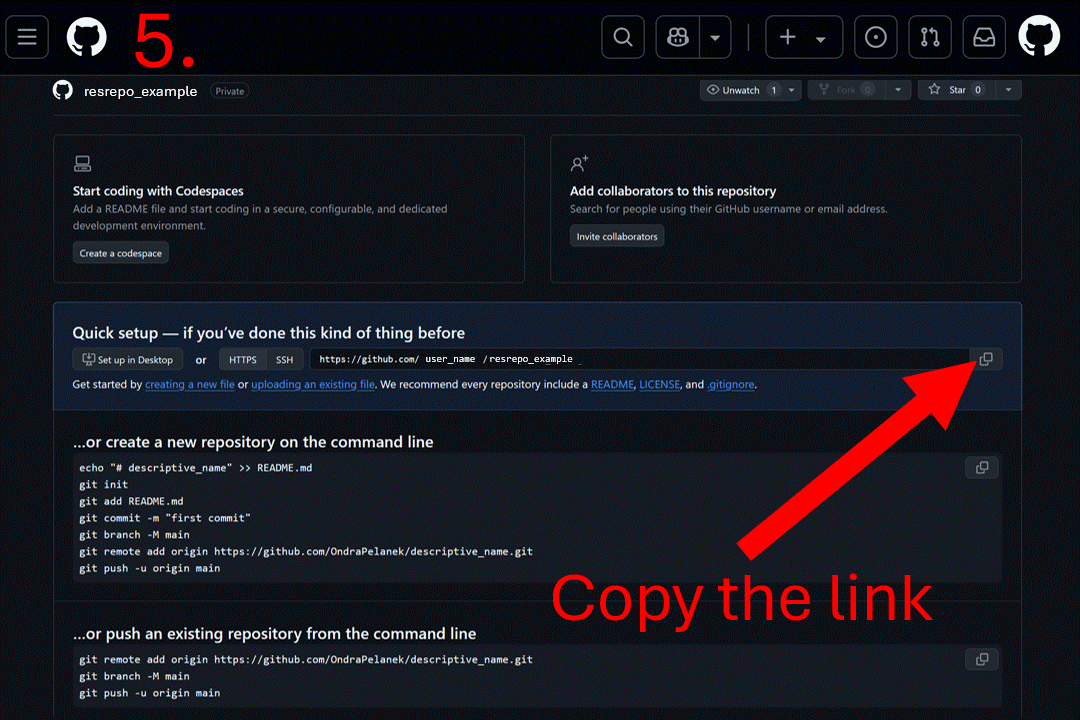
Then, you can clone the repository on your computer. You can do this in RStudio by clicking on the File tab, then New Project…, Version Control, Git, and pasting the URL of your repository.
We will now use resrepo to initialise the repository.
First, make sure that your working directory is set within the
git repository:
getwd()
#> [1] "/tmp/RtmpM3RrgE/resrepo_example"We can now initialise the repository:
library(resrepo)
init_resrepo()
#> [1] TRUELet us look at the content of our new repository:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ └── README.md
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ │ └── README.md
#> │ └── raw
#> │ ├── README.md
#> │ └── original
#> ├── results
#> │ └── README.md
#> └── writing
#> └── README.mdYou should now modify the main README.md, found at the root of the project, to describe your project. That document will act as the landing page of your git repository in GitHub/GitLab.
Similar tools
Scientific projects contain both code and data: git is
designed to manage software code, but it is not suited to track large
data files. There are extensions of git, such as
git-lfs and git-annex that can handle data,
but they can be complex to set up and difficult to use, especially when
sharing your repository among collaborators. resrepo
encourages good habits to manage your data alongside your code in plain
git, making it easy to share your project with
collaborators, thus ensuring reproducible science and a tidy repository
that can used for publication of your work.
A step by step example
We will now illustrate how to populate a simple project with
resrespo.
In this project, we have gone to the field and measured several
individuals of a new imaginary species of penguin called Tux. We have
collected data on their bill length and depth, flipper length, body mass
and sex. We will store this data, of which we have a copy in the
package, as a CSV file named “tux_measurements.csv” in the
data/raw/original directory:
file.copy(
from = system.file("vignette_example/tux_measurements.csv",
package = "resrepo"
),
to = path_resrepo("/data/raw/original/tux_measurements.csv"),
overwrite = TRUE
)
#> [1] TRUELet’s have a look at the repository:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ └── README.md
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ │ └── README.md
#> │ └── raw
#> │ ├── README.md
#> │ └── original
#> │ └── tux_measurements.csv
#> ├── results
#> │ └── README.md
#> └── writing
#> └── README.mdWe will then write a script to download data on three other species
of penguins from the palmerpenguins dataset. We will call
this script s01_download_penguins.Rmd, and store the data
in data/raw/s01_download_penguins.
resrepo can generate a pre-edited Rmd file for you to
use as a template for your analysis . This is better than the default
template in RStudio, and we will explore some of its additional features
further below. To create a new Rmd file from the resrepo
template Rmd file, use the create_rmd function and specify
the file name and which folder it should be in (you do not need to
include the .Rmd file extension).
create_rmd("code/s01_download_penguins")We can replace the explanatory text and examples in this Rmd with only a simple chunk of code:
library(resrepo)
# define the output directory (named after the script) and create it
output_dir <- path_resrepo("/data/raw/s01_download_penguins")
if (!dir.exists(output_dir)) {
dir.create(output_dir, recursive = TRUE)
}
# load the data
data("penguins", package = "palmerpenguins")
# write the data
write.csv(
x = penguins, file = file.path(output_dir, "palmer_penguins.csv"),
row.names = FALSE
)Note that, to set the output directory, we used the function
path_resrepo, which allows us to use paths relative to the
root of the git repository, irrespective of where the scripts are.
Alternatively, you could use a relative path from the script,
e.g. ../data/raw/s01_download_penguins. The ..
means “go up one directory”, so from the code directory, it
would go to the root of the repository, then into
data/raw/s01_download_penguins. If you want more
information on relative paths, have a look at this link.
Add the chunk above to our s01_download_penguins.Rmd and save it.
We can see the script in the code directory:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ └── s01_download_penguins.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ │ └── README.md
#> │ └── raw
#> │ ├── README.md
#> │ └── original
#> │ └── tux_measurements.csv
#> ├── results
#> │ └── README.md
#> └── writing
#> └── README.mdYou can now open the Rmd and knit it to download the data.
#> processing file: s01_download_penguins.Rmd
#> output file: s01_download_penguins.knit.md
#>
#> Output created: s01_download_penguins.pdfWe can see that the data was saved in the right place:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ └── s01_download_penguins.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ │ └── README.md
#> │ └── raw
#> │ ├── README.md
#> │ ├── original
#> │ │ └── tux_measurements.csv
#> │ └── s01_download_penguins
#> │ ├── palmer_penguins.csv
#> │ └── renv_s01_download_penguins.json
#> ├── results
#> │ ├── README.md
#> │ └── s01_download_penguins
#> │ ├── README.md
#> │ └── s01_download_penguins.pdf
#> └── writing
#> └── README.mdNote that knitting our Rmd also created the new directory
results/s01_download_penguins to store the knitted
markdown. Standard Rmd documents save the outputs in the same directory
as the code, which would break the rule of separation of code and
outputs. To avoid this, rmd documents created with
resrepo::create_rmd(), have their knitted output stored in
the right place in results by default. In this case, this document is
pretty minimal (it just repeats the code used to download and save), but
later on we will use it to document our analysis.
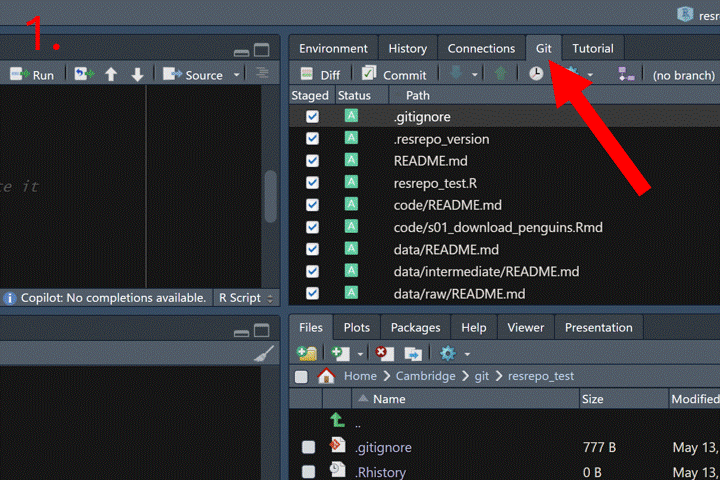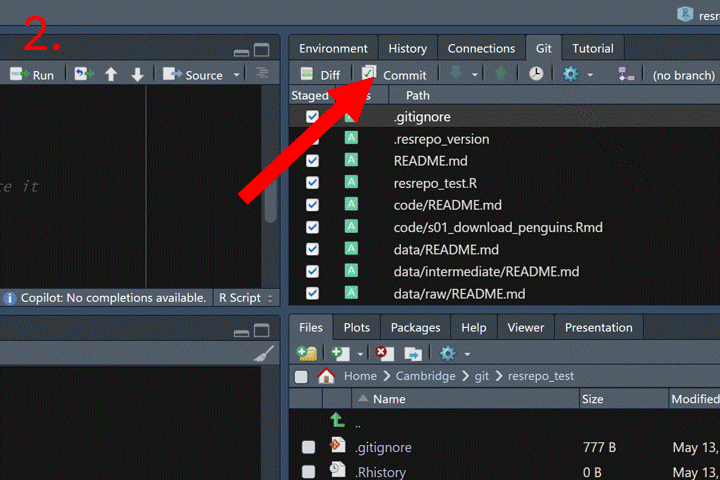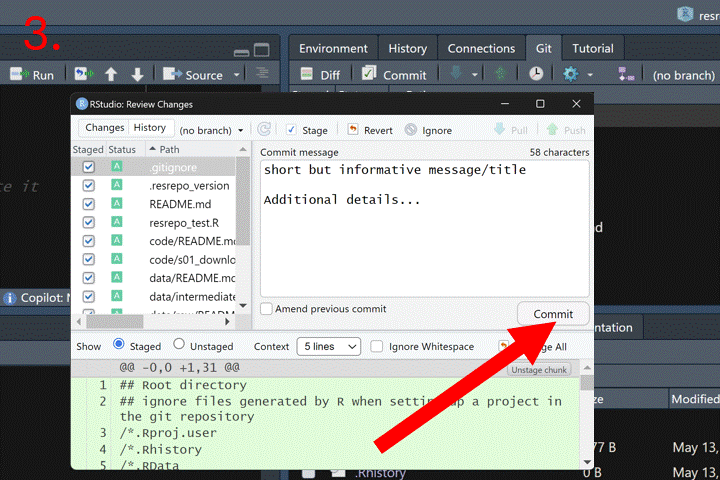
Now that we have new files (the code to download and the resulting data and output), we should commit our changes to the repository. In RStudio, open the Git tab on the right-hand side, click the Commit button, write a descriptive commit message, and then click Commit to finalize.
We now want to merge our data on the new species with the reference
dataset, and clean the data (e.g. remove missing data). We will copy
over an Rmd from the package, which was originally created with
resrepo::create_rmd:
file.copy(
from = system.file("vignette_example/s02_merge_clean.Rmd",
package = "resrepo"
),
to = path_resrepo("/code/s02_merge_clean.Rmd"),
overwrite = TRUE
)
#> [1] TRUEWe can check that the script is in the right place:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ ├── s01_download_penguins.Rmd
#> │ └── s02_merge_clean.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ │ └── README.md
#> │ └── raw
#> │ ├── README.md
#> │ ├── original
#> │ │ └── tux_measurements.csv
#> │ └── s01_download_penguins
#> │ ├── palmer_penguins.csv
#> │ └── renv_s01_download_penguins.json
#> ├── results
#> │ ├── README.md
#> │ └── s01_download_penguins
#> │ ├── README.md
#> │ └── s01_download_penguins.pdf
#> └── writing
#> └── README.mdHave a look at this Rmd document. Note how we set all input paths at the very beginning of a script, so that it is easy to know what is needed to run that script. This habit makes it much easier to share the script with others.
You can now knit the Rmd to merge the data and clean it.
#> processing file: s02_merge_clean.Rmd
#> output file: s02_merge_clean.knit.md
#>
#> Output created: s02_merge_clean.pdfWe can see that the data was saved in the correct
/data/intermediate subdirectory:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ ├── s01_download_penguins.Rmd
#> │ └── s02_merge_clean.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean
#> │ │ ├── penguins_na_omit.csv
#> │ │ └── renv_s02_merge_clean.json
#> │ └── raw
#> │ ├── README.md
#> │ ├── original
#> │ │ └── tux_measurements.csv
#> │ └── s01_download_penguins
#> │ ├── palmer_penguins.csv
#> │ └── renv_s01_download_penguins.json
#> ├── results
#> │ ├── README.md
#> │ ├── s01_download_penguins
#> │ │ ├── README.md
#> │ │ └── s01_download_penguins.pdf
#> │ └── s02_merge_clean
#> │ ├── README.md
#> │ └── s02_merge_clean.pdf
#> └── writing
#> └── README.mdAgain, note that a markdown output was placed in an appropriate
subdirectory of results.
We are now ready to do some analysis. We will start with a PCA, with the aim of testing whether our tux penguin is clearly distinct from other penguin species. We use a script from the package, which we copy over to our repository:
file.copy(
from = system.file("vignette_example/s03_pca.Rmd", package = "resrepo"),
to = path_resrepo("/code/s03_pca.Rmd"),
overwrite = TRUE
)
#> [1] TRUEOpen it. Again, note our input and output paths are all set at the top of the script, to make it easy to understand what is needed to run the script. You should now run (knit) your script.
Let’s check the repository once again:
fs::dir_tree()
#> .
#> ├── README.md
#> ├── code
#> │ ├── README.md
#> │ ├── s01_download_penguins.Rmd
#> │ ├── s02_merge_clean.Rmd
#> │ └── s03_pca.Rmd
#> ├── data
#> │ ├── README.md
#> │ ├── intermediate
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean
#> │ │ ├── penguins_na_omit.csv
#> │ │ ├── renv_s02_merge_clean.json
#> │ │ └── renv_s03_pca.json
#> │ └── raw
#> │ ├── README.md
#> │ ├── original
#> │ │ └── tux_measurements.csv
#> │ └── s01_download_penguins
#> │ ├── palmer_penguins.csv
#> │ └── renv_s01_download_penguins.json
#> ├── results
#> │ ├── README.md
#> │ ├── s01_download_penguins
#> │ │ ├── README.md
#> │ │ └── s01_download_penguins.pdf
#> │ ├── s02_merge_clean
#> │ │ ├── README.md
#> │ │ └── s02_merge_clean.pdf
#> │ └── s03_pca
#> │ ├── README.md
#> │ ├── s03_pca.pdf
#> │ └── s03_pca_files
#> │ └── figure-latex
#> │ └── pca_plot-1.png
#> └── writing
#> └── README.mdNow you just need to commit your changes as you did above, and we have a full repository with data, code and results, with a clear structure that should be easy to interpret.
Some tips on how to use GitHub
Commit Think carefully about your commit messages and branch names, as they will be very useful for returning to past changes for both you and others (for example when a project is made publicly available). Please make them informative.
Merge Once you merge a branch into master, kill the branch to avoid problems in the future. Do not name a branch with the name of a branch that has been merged/deleted recently, as this may create problems.
Binary files Do not upload binary data (e.g.*.rds): changes in such files are too heavy to be handled by GitHub.
GitHub setup
If you already have a ssh key, follow these steps: https://docs.github.com/en/authentication/connecting-to-github-with-ssh/checking-for-existing-ssh-keys
If you need to generate a new key and add it to GitHub: 1. Generate ssh key Source: https://docs.github.com/en/authentication/connecting-to-github-with-ssh/generating-a-new-ssh-key-and-adding-it-to-the-ssh-agent
2. add key to account Source: https://docs.github.com/en/authentication/connecting-to-github-with-ssh/adding-a-new-ssh-key-to-your-github-account
3. Test connection Source: https://docs.github.com/cn/authentication/connecting-to-github-with-ssh/testing-your-ssh-connection